Binning a Metagenomic Assembly¶
A common approach following metagenome assembly is binning, a process by which assembled contigs are collected into groups or ‘bins’ that might then be assigned some taxonomic affiliation. There are many different tools that can be used for binning (see CAMI review for more details). Here, we will be using MaxBin and MetaBAT, which are both user friendly and highly cited. To use these binners, we will first need to map our data against the assembled metagenome using bwa and then estimate relative abundances by contig. We will then inspect the bins generated by MaxBin and MetaBAT using VizBin.
Installing binners¶
MaxBin
cd
curl https://downloads.jbei.org/data/microbial_communities/MaxBin/getfile.php?MaxBin-2.2.2.tar.gz > MaxBin-2.2.2.tar.gz
tar xzvf MaxBin-2.2.2.tar.gz
cd MaxBin-2.2.2
cd src
make
cd ..
./autobuild_auxiliary
export PATH=$PATH:~/MaxBin-2.2.2
MetaBAT
cd
curl -L https://bitbucket.org/berkeleylab/metabat/downloads/metabat-static-binary-linux-x64_v0.32.4.tar.gz > metabatv0.32.4.tar.gz
tar xvf metabatv0.32.4.tar.gz
Time to finally run the Binners! Note: MaxBin can take a lot of time to run and bin your metagenome. As this is a workshop, we are doing three things that sacrifice quality for speed.
- We are only using 2 of the 6 datasets that were generated for the this project. Most binning software, rely upon many samples to accurately bin data. And, we have subsampled the data to make it faster to proess.
- We did quick and dirty contig estimation using samtools. If you use MaxBin, I would recommend allowing them to generate the abudance tables using Bowtie2. But, again, this will add to the run time.
- We are limiting the number of iterations that are performed through the MaxBin expectation-maximization algorithm (5 iterations instead of 50+). This will likely limit the quality of the bins we get out. So, users beware and read the user’s manual before proceeding with your own data analysis.
Get a rough count of the total number of reads mapping¶
Q. Why is the read counts per contigs not an accurate number?
cd ~/mapping
for i in *sorted.bam
do
samtools idxstats $i > $i.idxstats
cut -f1,3 $i.idxstats > $i.counts
done
Binning 1 - MaxBin¶
–
Maxbin uses read coverage & tetranucleotide frequencies for each contig, and marker gene counts for each bin
First, we will get a list of the count files that we have to pass to MaxBin
mkdir ~/mapping/binning
cd ~/mapping/binning
ls ../*counts > abundance.list
Now, on to the actual binning
run_MaxBin.pl -contig ../subset_assembly.fa -abund_list abundance.list -max_iteration 5 -out mbin
This will generate a series of files. Take a look at the files generated. In particular you should see a series of *.fasta files preceeded by numbers. These are the different genome bins predicted by MaxBin.
Take a look at the mbin.summary file. What is shown?
Now, we are going to generate a concatenated file that contains all of our genome bins put together. We will change the fasta header name to include the bin number so that we can tell them apart later.
for file in mbin.*.fasta
do
num=${file//[!0-9]/}
sed -e "/^>/ s/$/ ${num}/" mbin.$num.fasta >> maxbin_binned.concat.fasta
done
And finally make an annotation file for visualization
echo label > maxbin_annotation.list
grep ">" maxbin_binned.concat.fasta |cut -f2 -d ' '>> maxbin_annotation.list
Binning 2 - MetaBAT¶
–
MetaBAT uses read coverage, coverage variance, & tetranucleotide frequencies for each contig. This is done with a custom script
cd ~/mapping
~/metabat/jgi_summarize_bam_contig_depths --outputDepth depth_var.txt *sorted.bam
Setup another “binning” directory for this tool’s results
mkdir ~/mapping/binning_metabat
cd ~/mapping/binning_metabat
Run MetaBAT script
Note that we are outputting info to a logfile
~/metabat/metabat -i ../subset_assembly.fa -a ../depth_var.txt --verysensitive -o metabat -v > log.txt
Make the .fasta file of all binned sequences
for file in metabat.*.fa
do
num=${file//[!0-9]/}
sed -e "/^>/ s/$/ ${num}/" metabat.$num.fa >> metabat_binned.concat.fasta
done
Make an annotation file of the bin numbers for annotation in VizBin
echo label > metabat_annotation.list
grep ">" metabat_binned.concat.fasta |cut -f2 -d ' '>> metabat_annotation.list
Visualizing the bins¶
Now that we have our binned data from both MetaBAT and MaxBin there are several different things we can do. One thing we might want to do is check the quality of the binning– a useful tool for this is CheckM. Today, for the sake of time, we will visualize the bins that we just generated using VizBin.
First, install VizBin::
cd
sudo apt-get install libatlas3-base libopenblas-base default-jre
curl -L https://github.com/claczny/VizBin/blob/master/VizBin-dist.jar?raw=true > VizBin-dist.jar
VizBin can run in OSX or Linux but is very hard to install on Windows. To simplify things we are going to run VizBin in the desktop emulator through JetStream (which is ... a bit clunky). So, go back to the Jetstream and open up the web desktop simulator.
Open the terminal through the desktop simulator and open VizBin:
java -jar VizBin-dist.jar
This should prompt VizBin to open in another window. First we will look at the output of the MaxBin assembly. Click the choose button to open file browser to navigate to the binning folder (~/mapping/binning). There you will find the concatenated binned fasta file (maxbin_binned.concat.fasta). Upload this file and hit run.
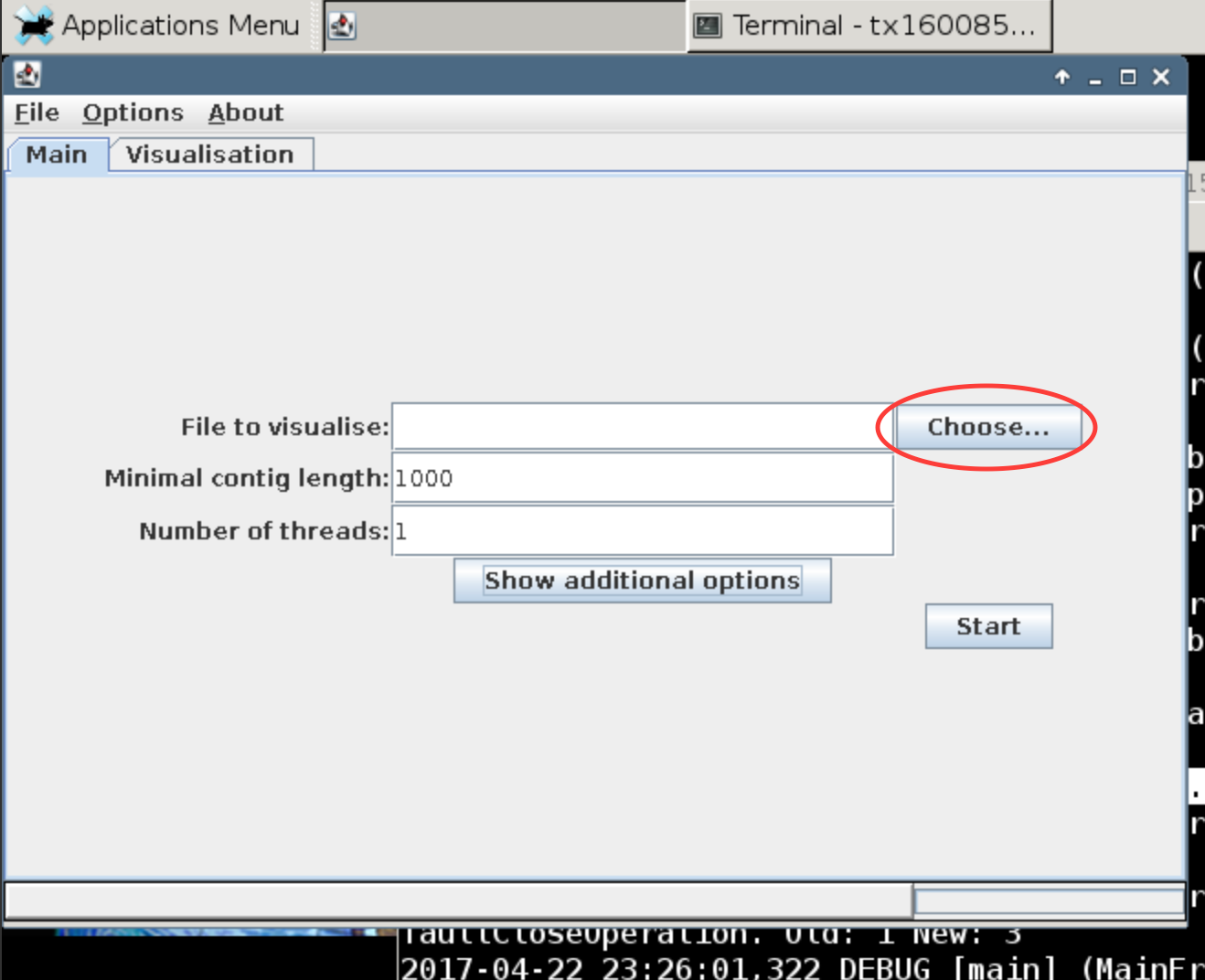
What do you see? Read up a bit on VizBin to see how the visualization is generated.
Now, upload the maxbin_annotation.list file as an annotation file to VizBin. The annotation file contains the bin id for each of the contigs in the assembly that were binned.
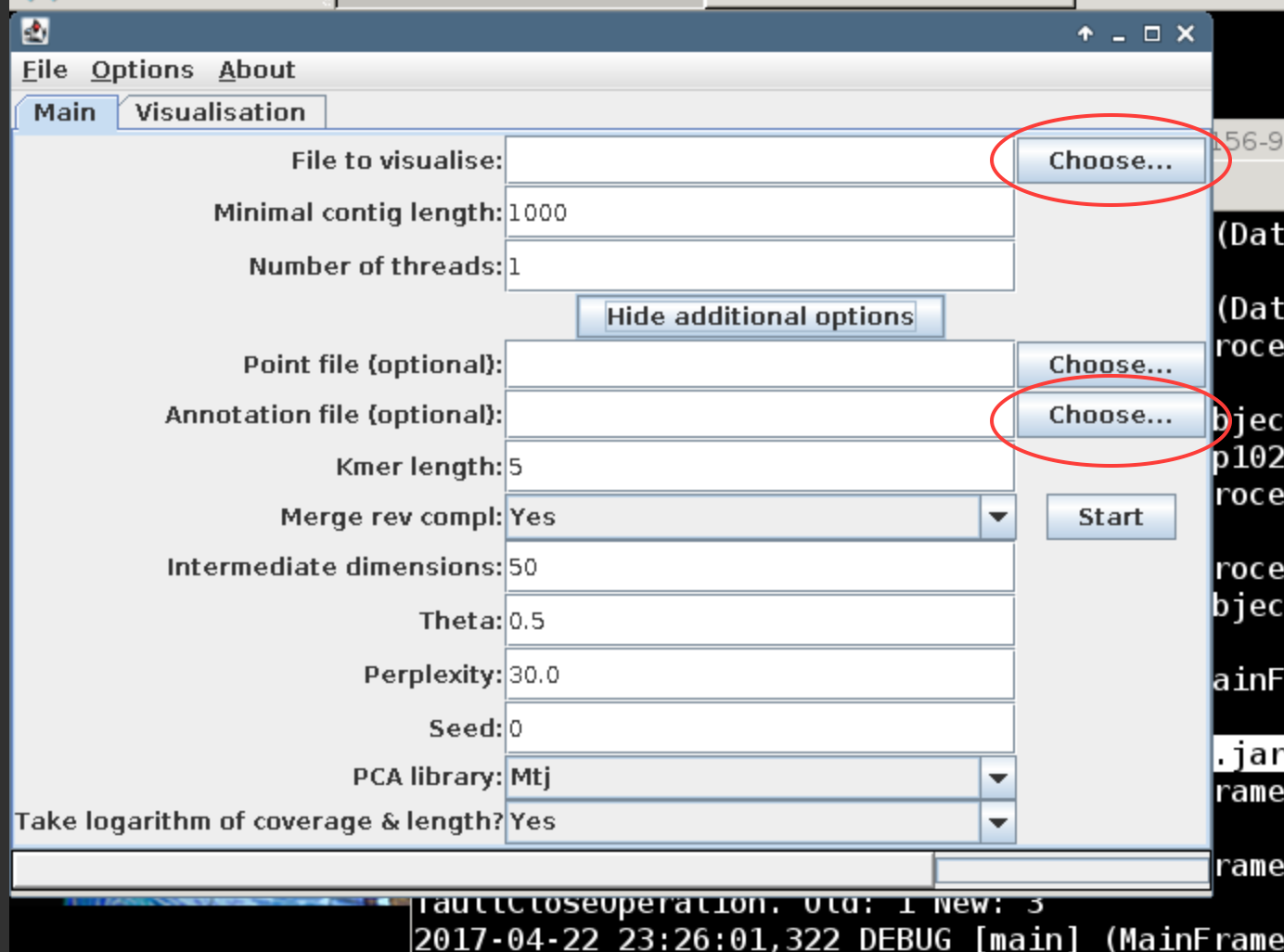
Now, do the same for MetaBat!
Compare the results of the two binning methods-
- How many bins were found?
- How distinct are the bins?
LICENSE: This documentation and all textual/graphic site content is released under Creative Commons - 0 (CC0) -- fork @ github.